Alexis E. Rose, Christopher D. Warner
Introduction
The health and performance of the gut is influenced by a myriad of factors including diet, genetics, environment, stress, intestinal barrier, immune status, and the microbiome. The alteration of one or more of these factors can lead to enteropathy, which is characterized as an increase in intestinal permeability, impairment of gut immune function, and nutrient malabsorption¹. An imbalance in the gastrointestinal tract can lead to deteriorating intestinal barrier function allowing for the translocation of pathogens and induce circulating immune regulators, thus generating a cyclical cause and effect relationship of increased inflammation and aberrant cytokine production². The positive feedback loop of inflammation in the gut can ultimately cause chronic disease states such as IBS-D, IBD, or HIV-associated enteropathy. Therefore, restoring balance within the GI tract is critical to overall health.
Immunoglobulins have a positive role in gut homeostasis. Serum-derived bovine immunoglobulin/protein isolate (SBI) is a spray-dried protein powder specially formulated to enrich the immunoglobulin content to help manage loose and frequent stools associated with gastrointestinal enteropathies. It is manufactured by fractionating edible grade bovine plasma to contain 90% protein, with most of the total protein comprised of immunoglobulins: 60% IgG, 5% IgM, and 1% IgA. The high concentration of immunoglobulins has shown to bind and neutralize opportunistic pathogens by immune and steric exclusion mechanisms using an in vitro cell culture model³. This binding of antigens helps limit the stimulation of immune regulators that modulate chronic intestinal inflammation when these microbial components translocate into the lamina propria.
The goal of this study was to expand the list of antigens to which SBI binds by adapting the ELISA used by Detzel et al³ to demonstrate binding of IgG to lipopolysaccharide, lipid A, and Pam3CSK4 antigens. New relevant antigens of interest were chosen based on the market needs for binding of gram-negative bacteria components and mold mycotoxins.
Overview of Antigens
Heliobacter pylori
Heliobacter pylori (H. pylori) is a gram-negative, spiral shaped bacterium that colonizes the human gastric mucosa. It is estimated that over half the world’s population carries H. pylori. Infection can lead to chronic atrophic gastritis, peptic ulcers, or gastric adenocarcinoma. Clinically, cytotoxicity-associated immunodominant antigen (CagA) positive H. pylori strains are more pathogenic and lead to disease states. CagA is injected directly into gastric epithelial cells by H. pylori and negatively influences tight junctions, cell proliferation, and morphology4.
Candida Albicans
Candida albicans (C. albicans) is a commensal yeast found commonly in the human oral mucosa, gut, vagina, and skin. When found in a dysbiosis state, C. albicans become pathogenic causing superficial and systemic infections. C. albicans are dimorphic and highly adaptable to change in environment (pH, temperature, CO2, nutrients).
The agglutinin-like protein 3 (Als3) plays a diverse role in C. albicans infection. Als3 functions as an adhesin, mediating attachment to epithelial cells, endothelial cells, and matrix proteins. It also has an active role in biofilm production and helps with iron uptake5. Lastly, Als3 is an invasin, binding to host cells receptors like E- and N-cadherin to endocytose the host cell6.
Cytolethal distending toxin
Cytolethal distending toxins (CDT) are heterotrimeric toxins produced by gram-negative bacteria associated with gut inflammation and disease. CDT activity relies on three subunits: CdtA, CdtB, and CdtC. CdtA and CdtC bind to the cell surface while CdtB acts as a nuclease, damaging host cell DNA resulting in cell cycle arrest. The individual domains display little to no toxicity until associating into a trimer of two cell binding domains and one CdtB. Cells infected with CDT display cytoplasm distention toxicity which gives the toxin its name7.
Shiga Toxin
Shiga toxin (Stx) is produced by both Shigella dysenteriae and some serogroups of Escherichia coli. Stx consist of two major subunits, A and B, with a single A subunit noncovalently binding to a pentamer of five identical B subunits. The subunits of the B pentamer bind to the cellular receptor, globotriaosylceramide, Gb3, found primarily on endothelial cells. Whereas the A subunit blocks protein synthesis by removing an adenine residue from the 28S rRNA of the 60S ribosome. Infection of Stx causes bloody diarrhea, abdominal pain, and is associated with Hemolytic Uremic Syndrome (HUS). Shiga toxin types1 is produced by E. coli but is virtually identical to Stx produced by S. dysenteriae, containing only a single amino acid difference in primary sequence8, 9.
Mycotoxins
Mycotoxins are secondary toxic metabolites produced by certain fungi. Aflatoxin M1 (AFM1) is produced by Aspergillus flavus and can be found in milk and other food products obtained from contaminated livestock. AFM1 is a metabolite of aflatoxin B1 (AFB1) which is found in maize and nuts and is known to cause liver cancer. AFM1 is possibly carcinogenic and is regulated by the US Food and Drug administration10.
Methods
Sample Preparation
A 10% SBI solution was prepared by dissolving 5 g of protein in 45 mL of deionized (DI) water. The solution was mixed for a minimum of 2 hours and centrifuged. Solutions were then filtered through a 0.45 µm syringe filter (Thermo Scientific, Waltham, MA). To prepare the denatured SBI control, a 10% solution was heated to 90°C, then allowed to cool. Denaturation was verified by low recovery of active IgG content by a turbidimetric immunoassay used for release testing ImmunoLin® product. The denatured protein control was then analyzed by dot blot to verify the ability of the detection antibody (sheep anti-bovine-HRP) to detect denatured IgG.
Materials
H. pylori lysate (MBS537291), CagA (MBS596143), agglutinin-like protein 3 (MBS1111464), Shiga like toxin-1 subunit B (MBS145496), cytolethal distending toxin subunit C (MBS1466408), and cytolethal distending toxin subunit A (MBS1308926) were purchased from MyBiosource, San Diego, CA. Candida albicans lysate (NAT41601-100) was purchased through CedarLane Labs, Burlington, NC. Aflatoxin M1-BSA (A6412) and carbonate-bicarbonate (C3401-100CAP) were purchased from Sigma, St. Louis, MO. Tris (T393), sodium chloride (S640), methanol (A412), sulfuric acid (SA200), and Corning clear polystyrene 96-well medium-binding microplates (07-200-98) were purchased from Fisher Scientific, Hampton, NH. PVDF transfer membrane (88518), Tween 20 (PI37538), Pierce Protein-Free (PBS) Blocking Buffer (37572), 3,3′, 5, 5′-tetramethylbenzidine (TMB) (N301), and SuperSignal Pico Chemiluminescent Substrate (34078) were purchased from Thermo Fisher Scientific, Waltham, MA. Sheep anti-bovine IgG-HRP (A10-118P) was purchased from Bethyl Laboratories, Montgomery, TX.
SBI-Antigen Binding: Dot Blot
A PVDF transfer membrane was activated in methanol for 30 seconds, washed, and transferred to water to equilibrate for 2 minutes. The membrane was then dotted with 5 µL of antigen (200, 100, 50 µg/mL) diluted in pH 7.6 TBS (50 mM Tris-Cl pH 7.5, 150 mM sodium chloride) and 0.05% Tween 20 (TBS-Tween). After the antigen was blotted, the membrane was allowed to dry before adding Blocking Buffer supplemented with 0.05% Tween 20. The membrane was allowed to block for one hour on a rocking platform at room temperature. After blocking, the membrane was washed 3 times with TBS-Tween for 5 minutes per wash. The membrane was then incubated for 1 hour at room temperature on a rocking platform in a 10% solution of SBI, diluted 1:100 in TBS-Tween, after which it was washed 5 times with TBS-Tween for 5 minutes per wash. The membrane was then incubated with 25 mL of 1:25,000 sheep anti-bovine IgG-HRP detection antibody in TBS-Tween for 1 hour at room temperature on the rocking platform and then washed 5 times for 5 minutes per wash. SuperSignal Pico Chemiluminescent substrate was added (4 mL of each kit reagent), and the membrane was scanned on the LICOR using the High Resolution Setting. The resulting chemiluminescent image was analyzed using ImageStudio.
SBI-Antigen Binding: ELISA
96-well medium binding plates were coated overnight at 4℃ with 100 µL of 5 µg/mL or 10 µg/mL antigen in 0.05 M carbonate-bicarbonate buffer, pH 9.6. Each well was washed 5 times with TBS-Tween. Plates were then blocked for 1 hour at room temperature using 300 µL Blocking Buffer and 0.05% Tween. Wells were washed again with TBS-Tween and then incubated with 100 µL SBI or denatured SBI in TBS-Tween for 1 hour at room temperature. Afterward, the wells were emptied and washed 5 times with TBS-Tween. Each well received 100 µL of sheep anti-bovine IgG-HRP added at 1:125,000 or 1:150,000 dilution in TBS-Tween and incubated for 1 hour at room temperature. Plates were again washed 5 times with TBS-Tween. The reaction was visualized by the addition of 100 µL of TMB substrate and stopped with the addition of 100 µL 0.2 M sulfuric acid. Absorbance was read at 450 nm using a microplate reader (Molecular Devices, Sunnyvale, CA).
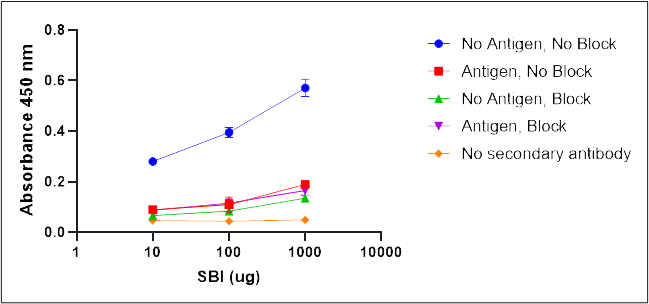
Control experiments were performed for each antigen specific ELISA to determine the non-specific binding of the detection antibodies and the specific recognition of the antigens by IgG contained in SBI. Non-specific binding of the detection antibodies was tested by running the ELISA without the SBI addition step. Blocking effectiveness was tested to demonstrate specificity of SBI IgGs for the test antigens by comparing the absorbance in unblocked wells to wells not coated with antigen. Functional binding of IgG for antigens was determined by comparing SBI treated wells to wells treated with a denatured SBI control. Overall binding specificity of SBI IgGs for the antigen was determined by a comparison of the complete ELISA with SBI to the denatured SBI control and the ELISA run without antigen but with blocking of the wells. The full list of controls is shown below.
- Binding control: antigen, no block, SBI, detection antibody, substrate
- Blocking control: no antigen, block, SBI, detection antibody, substrate
- Non-specific surface binding control: no antigen, no block, SBI, detection antibody, substrate
- Detection antibody background control: antigen, block, no SBI, detection antibody, substrate
- SBI background control: antigen, block, SBI, no detection antibody, substrate
- Functional binding control: antigen, block, denatured SBI, detection antibody, substrate
Results
Dot Blot
The binding of C. ablicans lysate, Als3, H. pylori lysate, CagA, Shiga-like toxin 1, aflatoxin M1, and CDT antigens by IgG in SBI were initially screened by a dot blot method. All antigens except the H. pylori lysate and aflatoxin M1 exhibited binding by dot blot. No additional testing was performed on the H. pylori lysate, but aflatoxin M1 antigen-IgG binding tested negative by ELISA (Appendix).
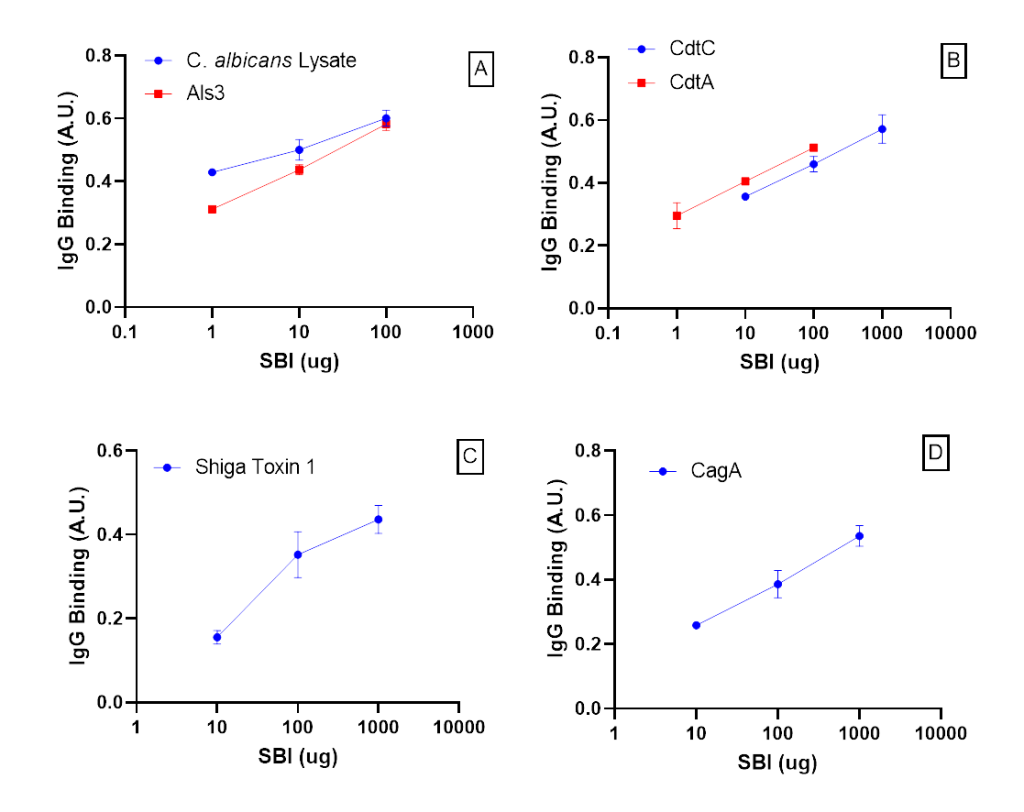
Figure 1. SBI binding affinity to various antigens A) SBI-IgG binds to C. albicans antigens, B) SBI-IgG binds to CDT antigens, C) SBI- IgG binds to Shiga-like toxin 1, D) SBI-IgG binds to a H. pylori CagA antigen. Error bars represent ± one standard deviation from triplicate data. Absorbance units (A.U.) measured at 450 nm.
ELISA
To demonstrate specificity of IgG binding, antigens were tested using a modified ELISA demonstrated in (Fig. 1, A-D). Absorbance of IgG bound to immobilized antigens increased proportionally to SBI concentration. SBI IgG specifically binds C. albicans lysate, C. albicans Als3 protein, CDT subunit A, CDT subunit B, Shiga-like toxin 1, and H. pylori CagA protein.
Specificity of immunoglobulin binding to each antigen was assessed by comparing the “Complete ELISA” curves shown in (Fig. 2, A-F) to control curves which excluded steps throughout the modified ELISA. SBI IgG binds specifically to antigens (Complete ELISA) when compared to the binding controls (Fig. 2, A-F). The complete ELISA displayed a higher absorbance compared to the blocking control (No antigen, Block), demonstrating IgG specificity for the antigen. Effective blocking is demonstrated by comparing wells treated with blocking buffer (No antigen, Block) to wells without blocking buffer (No antigen, No block). In summary, results from the specificity and binding controls indicate IgG binds specifically to each antigen in Table 1.
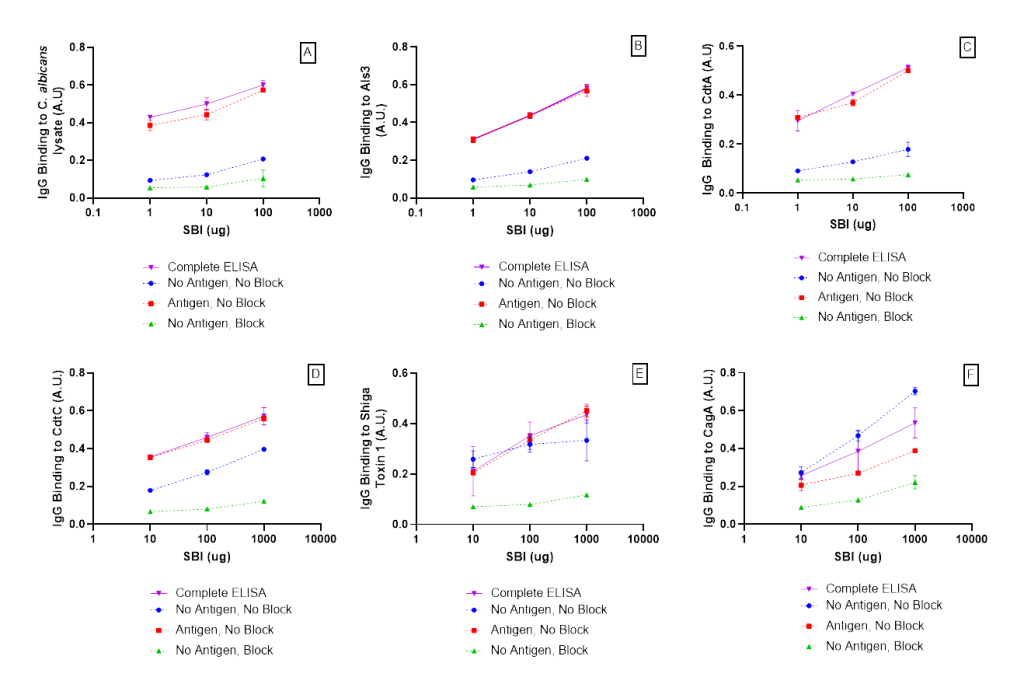
Figure 2. SBI IgG had a higher affinity for antigen protein (Complete ELISA) when compared to the blocking protein (-antigen, +block). Effective blocking is demonstrated by comparing wells treated with blocking buffer (-antigen. +block) to wells not treated with blocking buffer (-antigen, – block) A) SBI IgG binding to C. Albicans, B) SBI IgG binding to Als3, C) SBI IgG binding to CdtA, D) SBI IgG binding to CdtC, E) SBI IgG binding to Shiga Toxin 1, F) SBI binding to CagA. Error bars represent ± one standard deviation from triplicate data. Absorbance units (A.U.) measure at 450 nm.
Functional binding of SBI was assessed by comparing the complete ELISA curves shown in (Fig. 3, A-F) to the denatured SBI control, which displayed no binding activity for each ELISA. The background controls, no addition of the detection antibody or SBI, showed no significant absorbance compared to the Complete ELISA curve.

Figure 3. Specificity control experiment IgG binding to A) C. albicans lysate, B) C. albicans Als3 antigen, C) CdtA. D) CdtC, E) Shiga-like toxin 1, and F) H. pylori CagA antigen. Error bars represent ± one standard deviation from triplicate data. Absorbance units (A.U.) measured at 450 nm. Absorbance units (A.U) measured at 450 nm. Absorbance units (A.U) measured at 540 nm.
Discussion
The IgGs contained in SBI specifically bind C. albicans lysate and Als3 protein, CDT subunits A and C, Shiga-like toxin 1 subunit B, and H. pylori CagA protein as determined by ELISA. While the dot blots for H. pylori lysate and aflatoxin M1 showed no binding that does not prove a lack of SBI binding to those antigens. The lack of binding to H. pylori lysate conflicts with the work by Horgan et al have previously demonstrate binding of Immunolin IgGs to H. pylori lysate11. The discrepancy in the two sets of data is likely due to strain differences between the H. pylori lysates. With the strain in this study likely having less CagA expression or recovery as binding of IgG to the purified CagA antigen was demonstrated by ELISA. The case of the aflatoxin dot blot results presents a different challenge. Aflatoxins are small molecules which are present problems for a traditional sandwich ELISA format due to presentation issues to the IgG molecules resulting from steric hinderances. To overcome this issue, a conjugate of BSA and aflatoxin-M1 was bound to the PVDF membrane and used for an ELISA (data not shown) which showed no SBI IgG binding. It is still likely that SBI can bind some mycotoxins as BSA has been shown to bind to alternariol mycotoxin and BSA is present in SBI12.
Conclusion
The immunoglobulin content of SBI has previously been shown to bind to a variety of microbial antigens (e.g., LPS, flagellin, peptidoglycan, etc) associated with gastrointestinal disorders. To continue to understand the broad impact of SBI, new relevant antigens of interest were chosen to test for IgG binding. The results presented here suggest SBI can bind to antigenic components from negative-gram bacteria (C. albicans, H. pylori, S. dysenteriae, and E. coli) commonly associated with GI inflammation and disease.
References
- Zhang Y-J, Li S, Gan R-Y, Zhou T, Xu D-P, Li H-B. Impacts of Gut Bacteria on Human Health and Diseases. International journal of molecular sciences. 2015;16(4):7493-7519.
- Ghosh SS, Wang J, Yannie PJ, Ghosh S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. Journal of the Endocrine Society. 2020;4(2)doi:10.1210/jendso/bvz039
- Detzel CJ, Horgan A, Henderson AL, et al. Bovine immunoglobulin/protein isolate binds pro-inflammatory bacterial compounds and prevents immune activation in an intestinal co-culture model. PloS one. 2015;10(4):e0120278. doi:10.1371/journal.pone.0120278
- Alexander SM, Retnakumar RJ, Chouhan D, et al. Helicobacter pylori in Human Stomach: The Inconsistencies in Clinical Outcomes and the Probable Causes. Frontiers in microbiology. 2021;12:713955. doi:10.3389/fmicb.2021.713955
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. Feb 15 2013;4(2):119-28. doi:10.4161/viru.22913
- Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. Feb 2011;10(2):168-73. doi:10.1128/EC.00279-10
- Scuron MD, Boesze-Battaglia K, Dlakić M, Shenker BJ. The Cytolethal Distending Toxin Contributes to Microbial Virulence and Disease Pathogenesis by Acting As a Tri-Perditious Toxin. Frontiers in cellular and infection microbiology. 2016;6:168. doi:10.3389/fcimb.2016.00168
- Melton-Celsa AR. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiology spectrum. Aug 2014;2(4):Ehec-0024-2013. doi:10.1128/microbiolspec.EHEC-0024-2013
- Chan YS, Ng TB. Shiga toxins: from structure and mechanism to applications. Applied Microbiology and Biotechnology. 2016/02/01 2016;100(4):1597-1610. doi:10.1007/s00253-015-7236-3
- Saha Turna N, Wu F. Aflatoxin M1 in milk: A global occurrence, intake, & exposure assessment. Trends in Food Science & Technology. 2021/04/01/ 2021;110:183-192. doi:https://doi.org/10.1016/j.tifs.2021.01.093
- Horgan A, Maas K, Henderson A, Detzel C, Weaver E. Serum-derived bovine immunoglobulin/protein isolate binds to pathogen-associated molecular patterns. The FASEB Journal. 2014;28(Supplement No. 1):836.6.
- Fliszár-Nyúl E, Lemli B, Kunsági-Máté S, et al. Interaction of Mycotoxin Alternariol with Serum Albumin. International journal of molecular sciences. May 12 2019;20(9)doi:10.3390/ijms20092352
Appendix